by Jesselyn Brown and Rosie Kidman
Often in science, it is assumed most of the big discoveries happen out in the field, on glorious expeditions into the unknown. While this is an essential step in research, most of the discoveries are made afterwards, where researchers spend months to years compiling their collected data into a coherent, well curated paper for the scientific community. Such is the case for the Antarctic marine invertebrate samples collected for the Marine Omics’ group! Rosie and Jesselyn have been sampling tissue for genomic analyses and preserving specimens for the Museum of Tropical Queensland in JCU’s own Townsville (Bebegu Yumba) Campus; a far stretch in distance and weather from Antarctica!


Rosie (left) and Jesselyn (right) dissecting tissue from echinoderms collected from Heard and McDonald Islands
Each specimen has to be appropriately sampled for genomic analysis and preserved for the museum’s collection. Some of the jarred specimens will eventually be on display for the public, allowing visualization of the creatures living below the ice. However, the most critical part of the project comes from the tiny tissue samples collected before the specimens are preserved.
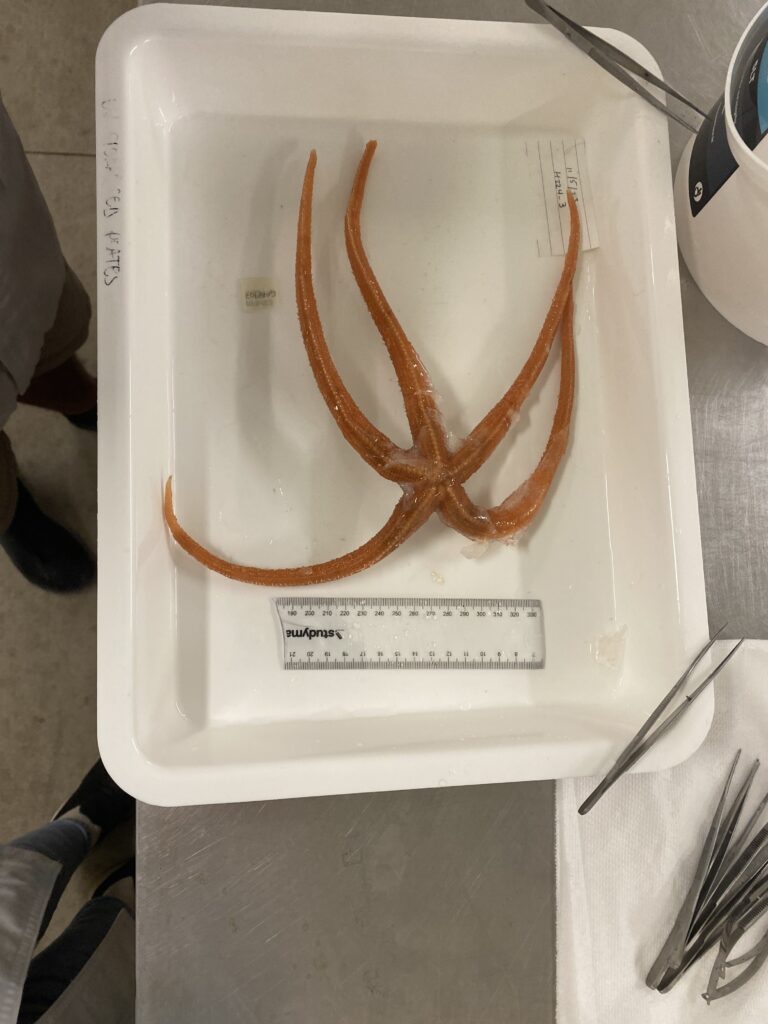

These echinoderms hold secrets about their evolutionary history and how it has been impacted by past climate and tectonic change.
For example, these echinoderms, as seen on Jesselyn’s dissection board, have tissue samples taken from their arms and from their tube feet. Although infinitesimally small, such DNA houses a wealth of information! This may reveal anything from population dynamics, evolutionary tracks and divergence records, survival rates over time, and occasionally, enable the prediction of species survival through a rapidly changing climate via paleorecords. These samples will play a vital role in piecing together genetic databases and allowing scientists to develop a deeper understanding of Antarctic fauna!