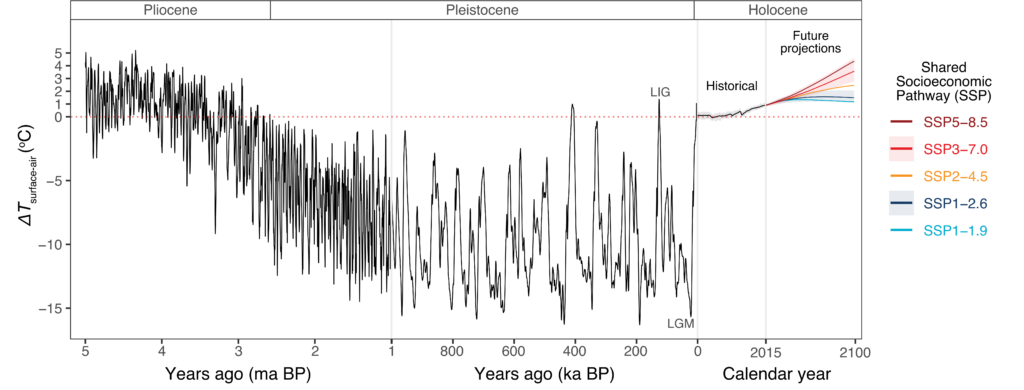
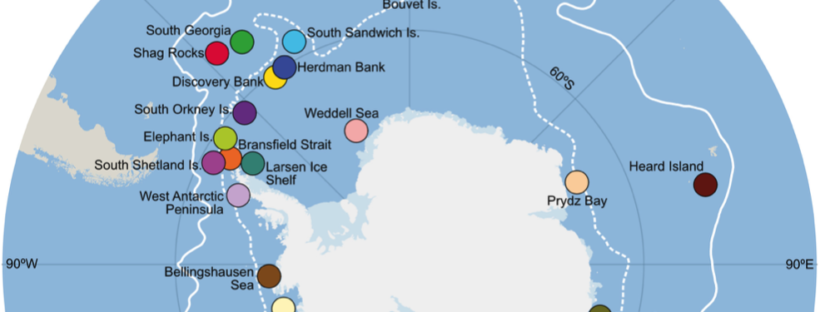
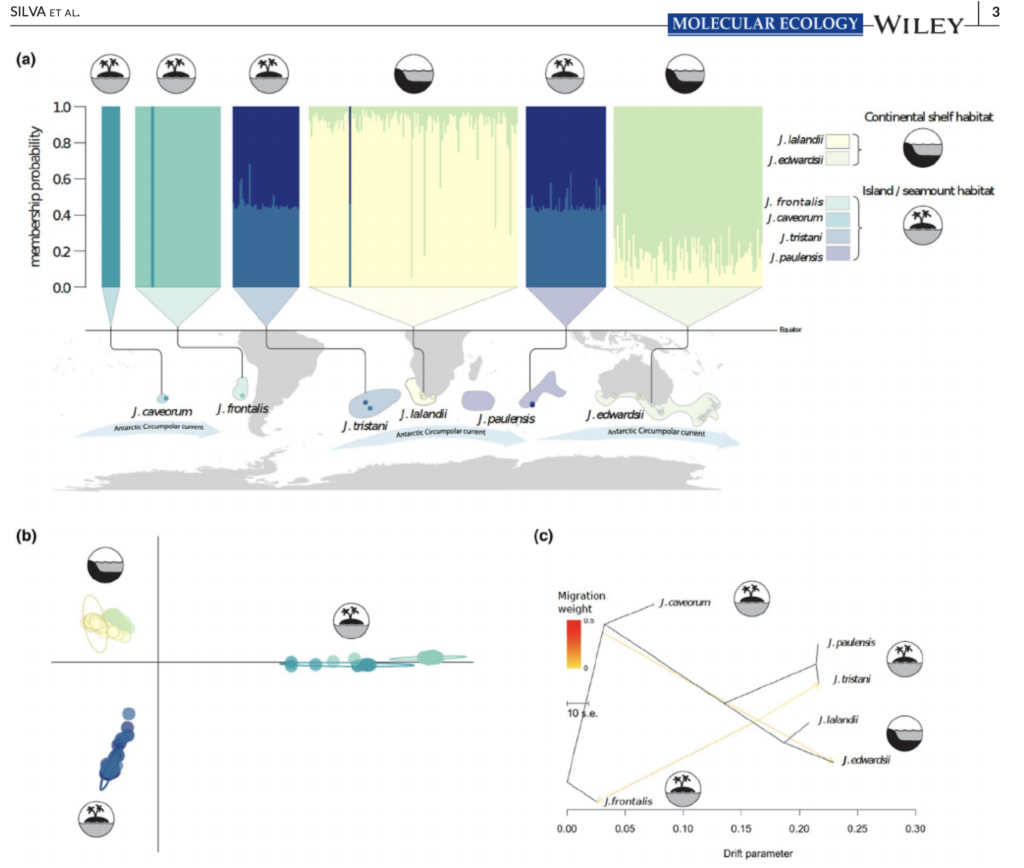
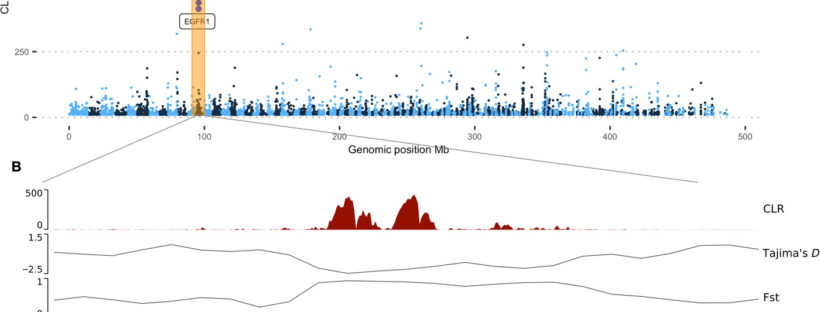
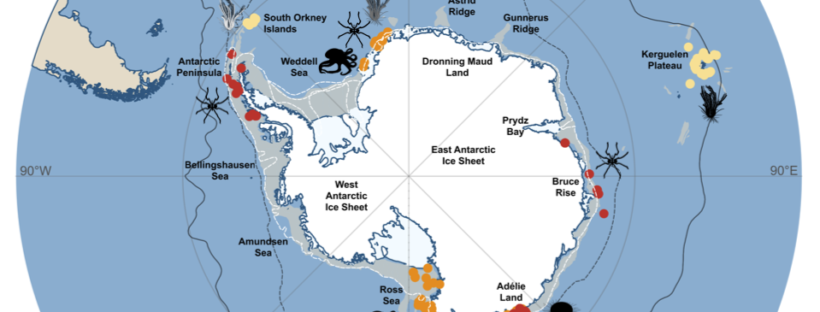
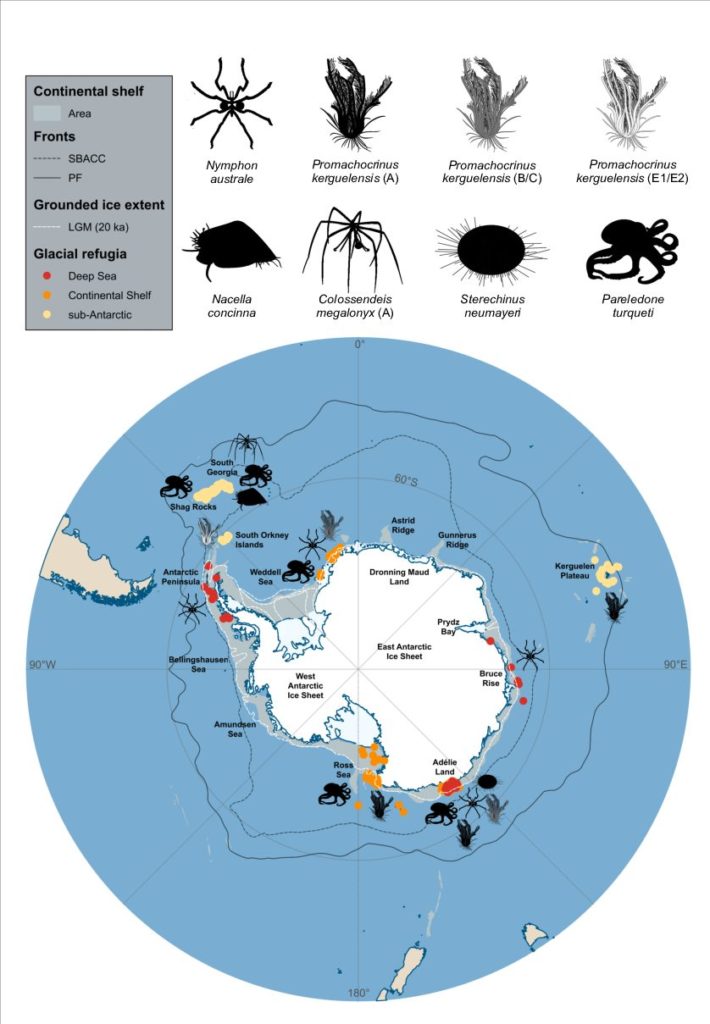
New editorial by Sally and Jan in Global Change Biology (full article here), explores the unprecedented environmental risks and consequences that the Southern Ocean is experiencing from current climate change. They also outlined that knowing how benthic fauna persisted through environmental extremes in the past may inform future predictions. Right now, understanding and preserving current genetic diversity and connectivity between populations will give species the best chance to adapt.